
Phylogenetic trees are visual representations of the evolutionary relationships between different organisms. They provide a way to understand how species are related to each other and can help scientists trace the history of life on Earth. Understanding how to read and interpret phylogenetic trees is an important skill for biologists, and the Phylogenetic Tree POGIL (Process Oriented Guided Inquiry Learning) activity is a valuable tool for teaching this concept.
The Phylogenetic Tree POGIL activity is designed to engage students in an active learning process, where they work collaboratively to answer questions and solve problems related to phylogenetic trees. By working in small groups and utilizing critical thinking, students are able to learn and retain information more effectively. The answer key for the Phylogenetic Tree POGIL provides students with the correct answers to the questions posed in the activity, allowing them to check their understanding and evaluate their progress.
The answer key for the Phylogenetic Tree POGIL can serve as a valuable resource for both students and instructors. For students, it provides a reference guide to ensure they are on the right track and understanding the material correctly. It can also help students identify any areas where they may be struggling and seek additional clarification from their instructor. For instructors, the answer key can be used as a teaching tool to highlight key concepts and guide class discussions.
What is a Phylogenetic Tree?

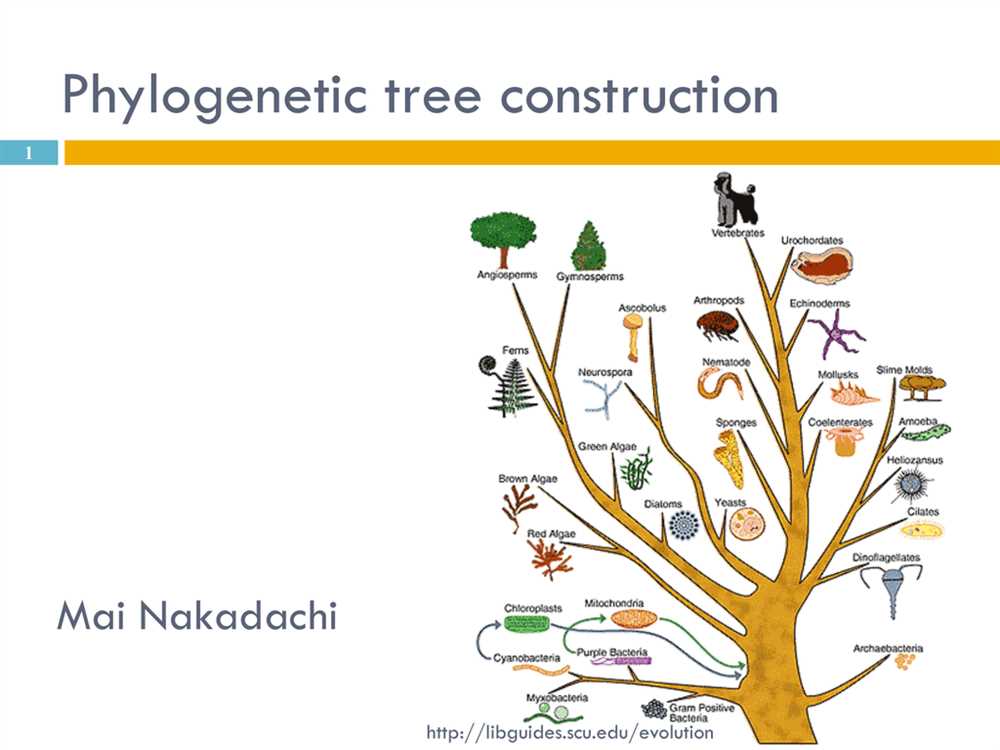
A phylogenetic tree, also known as an evolutionary tree or a tree of life, is a visual representation of the evolutionary relationships between different species. It is a branching diagram that shows the common ancestry and divergence of organisms over time.
The main purpose of a phylogenetic tree is to illustrate the evolutionary history and relationships of organisms, allowing scientists to understand the patterns of biological diversity and how different species are related to each other. It is a powerful tool for studying evolutionary biology and understanding the processes of speciation and adaptation.
A phylogenetic tree is constructed based on the similarities and differences between different species and their shared ancestors. The tree is built using various methods, such as comparing genetic sequences, morphological characteristics, or behavioral traits. By analyzing these data, scientists can determine the order and timing of evolutionary events, such as the emergence of new species or the extinction of ancient lineages.
The structure of a phylogenetic tree consists of branches, nodes, and leaves. The branches represent the evolutionary lineages, with the length of the branches indicating the amount of evolutionary change that has occurred. The nodes, or branching points, represent the common ancestors of different species, while the leaves represent the extant species or groups of species.
Importance of Phylogenetic Trees in Biology
Phylogenetic trees are an essential tool in biology for understanding the relationships between different species and their evolutionary history. These trees provide a visual representation of the evolutionary relationships based on genetic and morphological data, allowing scientists to understand how species are related to each other and how they have evolved over time.
One of the key benefits of phylogenetic trees is their ability to infer common ancestry. By comparing the similarities and differences in DNA sequences or physical traits, scientists can determine whether species share a common ancestor and how recently they diverged. This information can be crucial for understanding the origins of species and how they have adapted to their environments.
The construction of phylogenetic trees also allows scientists to make predictions about the characteristics and behaviors of species that have not yet been studied. By placing an unknown species within a phylogenetic tree and comparing its traits to those of closely related species, researchers can make educated guesses about its evolutionary history and likely traits. This can help guide further research and conservation efforts.
Phylogenetic trees are also important in areas such as medicine and agriculture. Understanding the evolutionary relationships between pathogens, for example, can help in the development of targeted treatments and vaccines. Similarly, in agriculture, phylogenetic trees can provide insights into the genetic relationships between different crop varieties, aiding in crop improvement and breeding programs.
In conclusion, phylogenetic trees are a crucial tool in biology for understanding evolutionary relationships and making predictions about species’ characteristics. They have wide-ranging applications in various fields and contribute to our knowledge of the natural world and its diversity.
Understanding the Components of a Phylogenetic Tree
A phylogenetic tree is a branching diagram that represents the evolutionary relationships among different species or groups of organisms. It is a visual representation of the evolutionary history, or phylogeny, of these species. Understanding the components of a phylogenetic tree is essential for interpreting and analyzing the information it conveys.
Branches: The branches on a phylogenetic tree represent the lineages of different species or groups of organisms. The length of the branches may vary, indicating the amount of evolutionary change that has occurred over time. Longer branches represent greater divergence and more evolutionary distance between species.
Nodes: Nodes, or branching points, on a phylogenetic tree represent common ancestors from which different species or groups of organisms have descended. The closer the nodes are to the root of the tree, the more ancient the common ancestor is. Nodes can be used to infer relationships between different groups of species and to estimate the timing of divergence events.
Root: The root of a phylogenetic tree represents the most recent common ancestor of all the species or groups of organisms included in the tree. It is the starting point from which the different branches and nodes diverge. The root is usually placed at the bottom of the tree, but in some cases, it may be drawn at the top.
Tips: When interpreting a phylogenetic tree, it is important to remember that the tree is a hypothesis based on available data. It represents the best current understanding of the relationships between different species, but new evidence and revisions may change the tree’s structure. Additionally, it is crucial to consider the underlying data and methods used to construct the tree, as different methods may yield different results.
In conclusion, a phylogenetic tree is a powerful tool for understanding the evolutionary relationships between different species or groups of organisms. By analyzing its components, such as branches, nodes, and the root, scientists can infer common ancestry, estimate divergence times, and gain insights into the patterns and processes of evolution.
Nodes and Branches
In a phylogenetic tree, nodes and branches play a crucial role in representing the evolutionary relationships among organisms. Node represents the common ancestor from which two or more lineages diverge, while branches depict the lineages themselves.
Nodes: Nodes are depicted as points or vertices on a phylogenetic tree. Each node represents a hypothetical common ancestor that gave rise to the lineages diverging from it. Nodes can be further classified into internal nodes and terminal nodes. Internal nodes represent ancestral species or hypothetical common ancestors that are no longer alive, while terminal nodes represent the living species or groups.
Branches: Branches, also known as edges, connect the nodes on a phylogenetic tree. They represent the evolutionary relationship between two lineages or groups. Each branch represents a lineage that has evolved from a common ancestor and can extend infinitely into the past or future. The length of a branch may represent the amount of genetic or evolutionary change that has occurred between two lineages or the time elapsed since they diverged. The angle between branches also indicates the relationship between different lineages.
The nodes and branches in a phylogenetic tree provide a visual representation of the evolutionary history and relatedness between different organisms or groups. They allow scientists to infer the genetic or evolutionary changes that have occurred over time and understand the patterns of speciation and diversification. By analyzing the nodes and branches, researchers can reconstruct the ancestral relationships and classify organisms into different taxa based on their shared evolutionary history.
Taxa and Clades

Taxa and clades are two important concepts in the field of phylogenetics, which is the study of the evolutionary relationships between organisms. A taxa refers to a group of organisms that share common characteristics and are classified together. These groups are hierarchical, with smaller groups nested within larger ones. For example, the domain is the highest level of classification, followed by kingdom, phylum, class, order, family, genus, and species. Each of these taxa represents a different level of relatedness between organisms.
Clades, on the other hand, are groups of organisms that are defined based on shared derived characteristics, or traits that are unique to the members of the clade and their common ancestor. Unlike taxa, which are defined based on a hierarchical classification system, clades are defined based on evolutionary relationships. This means that organisms within a clade share a more recent common ancestor with each other than with organisms outside of the clade.
For example, let’s consider the clade of mammals. This clade includes all organisms that are descended from a common ancestor that had hair and produced milk to nourish their young. Organisms within the mammal clade, such as humans and dogs, share these derived characteristics and are more closely related to each other than to organisms outside of the clade, such as birds or reptiles.
In phylogenetic trees, taxa are represented as branches or tips, while clades are represented as branches that include multiple taxa. These trees illustrate the evolutionary relationships between organisms, with branches that diverge from a common ancestor. Each branching point represents a speciation event, where a new species evolves from a common ancestor.
Understanding taxa and clades is crucial for phylogenetic analysis as it allows scientists to accurately classify and study organisms based on their evolutionary history. By identifying and comparing shared derived characteristics, researchers can reconstruct the evolutionary relationships between species and gain insights into their evolutionary history and biodiversity.
Constructing a Phylogenetic Tree
A phylogenetic tree is a visual representation of the evolutionary relationships between different organisms. It shows how species are related to each other through a common ancestor and helps scientists understand the patterns of evolution. Constructing a phylogenetic tree involves analyzing various characteristics and traits of organisms and using this information to determine their evolutionary history.
To construct a phylogenetic tree, scientists typically start by selecting a group of organisms that they want to study. These organisms can be closely related or from different taxonomic groups, depending on the research question. Once the organisms are selected, scientists gather data on their physical traits, genetic sequences, or other characteristics.
Comparative analysis is a crucial step in constructing a phylogenetic tree. Scientists compare the traits and characteristics of different organisms to identify similarities and differences. These shared traits can indicate a common ancestry between organisms. For example, if two species have similar anatomical structures, it suggests that they share a recent common ancestor.
Some key features that can be used for comparative analysis include:
- Homologous traits: Similar structures or traits that are derived from a common ancestor, such as the pentadactyl limb structure found in mammals and reptiles.
- Analogous traits: Similar structures or traits that have independently evolved in different lineages, often due to similar environmental pressures, such as the wings of bats and birds.
- Molecular data: Genetic sequences, such as DNA or RNA, can be analyzed to identify similarities and differences between organisms.
Once the comparative analysis is done, scientists use this information to construct a phylogenetic tree. The tree is typically represented as a branching diagram, where each branch represents a different species or group of species. The branches indicate the evolutionary relationships, with closer branches indicating more recent common ancestors.
Constructing a phylogenetic tree is an ongoing process, and new data and research can lead to revisions and updates. It is an essential tool in understanding the evolutionary history of life on Earth and helps scientists unravel the complexities of biodiversity.
Methods of Phylogenetic Tree Construction
Phylogenetic trees are used to represent the evolutionary relationships between different species. These trees can be constructed using various methods, each with its own advantages and limitations. One common method is the use of molecular data, specifically DNA or protein sequences, to infer the relationships between species. This method is based on the assumption that species with more similar sequences are more closely related.
To construct a phylogenetic tree using molecular data, researchers first need to obtain sequences from the species of interest. These sequences are then aligned to determine the similarities and differences between them. The aligned sequences are then used to build a phylogenetic tree using computational algorithms. These algorithms calculate the evolutionary distances between sequences and use this information to infer the branching patterns in the tree.
Another method used in phylogenetic tree construction is the analysis of morphological traits. This method involves comparing the physical characteristics of different species to determine their evolutionary relationships. Researchers identify specific traits that are shared between species and use this information to construct a phylogenetic tree. This method has been used for many years, particularly in the study of fossils where molecular data is not available.
In addition to molecular and morphological data, other types of data can also be used in phylogenetic tree construction. For example, researchers can use geographical and ecological data to infer relationships between species. These data can provide insights into how different species have evolved and adapted to different environments. By combining multiple types of data, researchers can obtain a more comprehensive understanding of the evolutionary relationships between species.
In conclusion, there are various methods available for constructing phylogenetic trees. The choice of method depends on the availability of data and the specific research question being addressed. Molecular data, morphological traits, and other types of data can all be used to infer evolutionary relationships between species and construct phylogenetic trees.